Poisson-truncated normal data
Table of Contents
Introduction
Zihao Wang shared an example simulated data set where ash gives a
surprising result.
Setup
import numpy as np import scipy.stats as st import scmodes
%matplotlib inline %config InlineBackend.figure_formats = set(['retina'])
import matplotlib.pyplot as plt plt.rcParams['figure.facecolor'] = 'w' plt.rcParams['font.family'] = 'Nimbus Sans'
Example
np.random.seed(1) N = 5000 lam = np.clip(np.random.normal(loc=10, scale=5, size=N), 0, None) x = np.random.poisson(lam=lam) s = np.ones(x.shape)
grid = np.linspace(x.min(), x.max(), 1000) F = st.norm(loc=10, scale=5) true_cdf = F.cdf(0) + F.sf(0) * F.cdf(grid)
cm = plt.get_cmap('Dark2') plt.clf() fig, ax = plt.subplots(2, 1) fig.set_size_inches(5, 4) ax[0].hist(x, bins=np.arange(x.max() + 1), color='k') ax[0].set_xlabel('$x$') ax[0].set_ylabel('$n_x$') for i, (method, t) in enumerate(zip(['zig', 'unimodal'], ['Point-Gamma', 'Unimodal'])): ax[1].plot(*getattr(scmodes.deconvolve, f'fit_{method}')(x, s), label=t, c=cm(i), lw=1) ax[1].plot(grid, true_cdf, label='Ground truth', c=cm(2), lw=1) ax[1].legend(frameon=False) ax[1].set_xlabel('$\lambda$') ax[1].set_ylabel('CDF') fig.tight_layout()
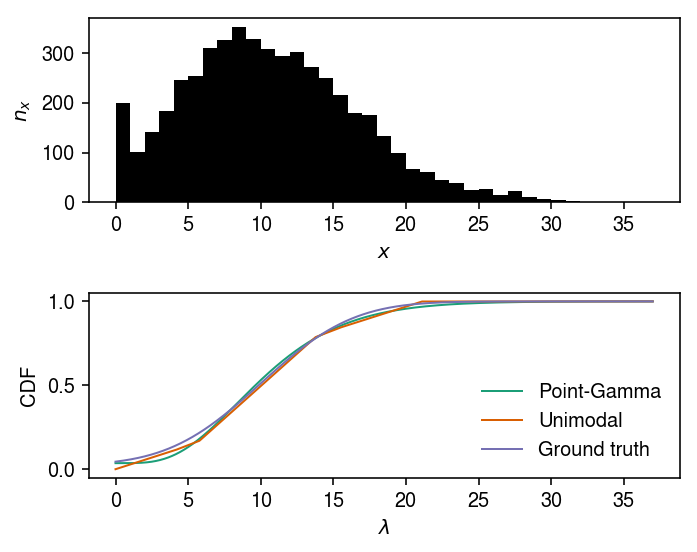
The grid we used to parameterize the unimodal distribution is:
np.exp(np.arange(np.log(1 / s.mean()), np.log((x / s).max()), step=.5 * np.log(2)))
array([ 1. , 1.41421356, 2. , 2.82842712, 4. , 5.65685425, 8. , 11.3137085 , 16. , 22.627417 , 32. ])