Pathological input to t-SNE
Table of Contents
Introduction
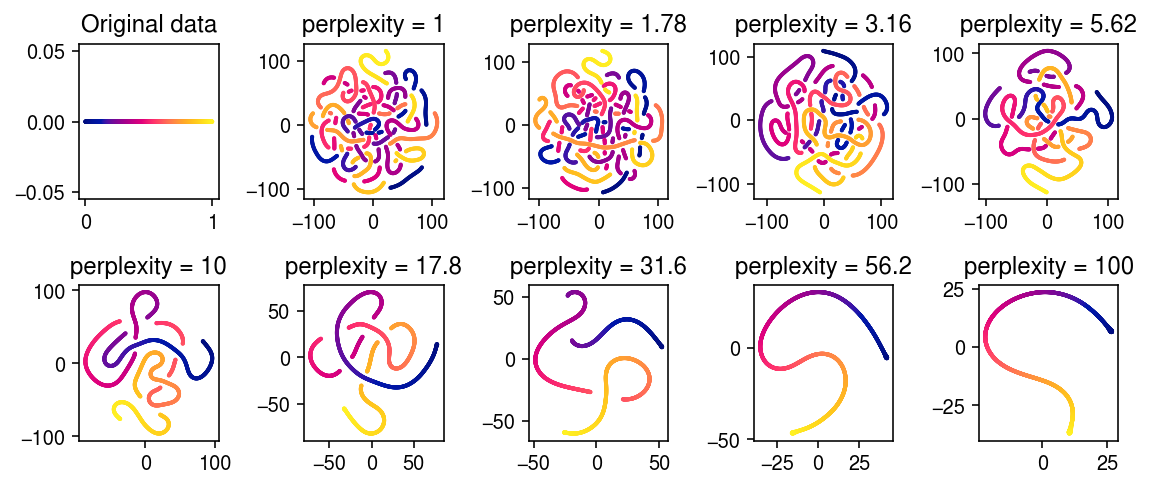
Peter Carbonetto reported an issue where t-SNE learns clusters from a topic model embedding where it shouldn’t. We give a simple example with continuously varying data where both t-SNE and UMAP invent clusters.
Setup
import numpy as np import sklearn.manifold as skm import umap
%matplotlib inline %config InlineBackend.figure_formats = set(['retina'])
import colorcet import matplotlib.pyplot as plt plt.rcParams['figure.facecolor'] = 'w' plt.rcParams['font.family'] = 'Nimbus Sans'
Example
Generate data varying in one dimension only
n = 2000 x = np.linspace(0, 1, n) c = colorcet.cm['bmy'](x) y = np.zeros(n) dat = np.vstack([x, y]).T
embeds = dict() for perp in np.logspace(0, 2, 9): print(f'fitting {perp:.3g}') # Important: seed the random initialization embeds[perp] = skm.TSNE(perplexity=perp, random_state=10).fit_transform(dat)
plt.clf() fig, ax = plt.subplots(2, 5) fig.set_size_inches(8, 3.5) ax[0,0].scatter(dat[:,0], dat[:,1], s=1, c=c) ax[0,0].set_title('Original data') for k, a in zip(embeds, ax.ravel()[1:]): temp = embeds[k] a.scatter(temp[:,0], temp[:,1], s=1, c=c) a.set_title(f'perplexity = {k:.3g}') fig.tight_layout()
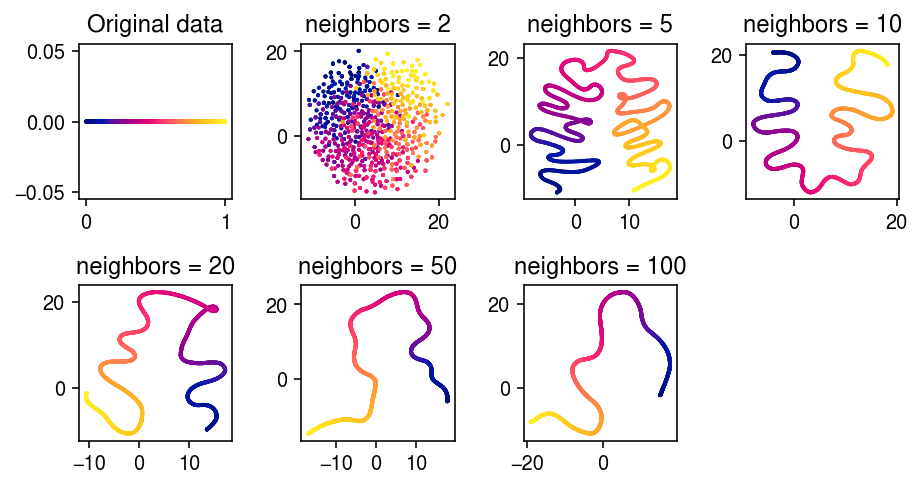
UMAP solves a different optimization problem. See what it does on this example.
umap_embeds = dict() for n_neighbors in (2, 5, 10, 20, 50, 100): print(f'fitting {n_neighbors}') # Important: seed the random initialization umap_embeds[n_neighbors] = umap.UMAP(n_neighbors=n_neighbors, min_dist=0, metric='euclidean', random_state=1).fit_transform(dat)
plt.clf() fig, ax = plt.subplots(2, 4) fig.set_size_inches(6.5, 3.5) ax[0,0].scatter(dat[:,0], dat[:,1], s=1, c=c) ax[0,0].set_title('Original data') for k, a in zip(umap_embeds, ax.ravel()[1:]): temp = umap_embeds[k] a.scatter(temp[:,0], temp[:,1], s=1, c=c) a.set_title(f'neighbors = {k}') ax[-1,-1].set_axis_off() fig.tight_layout()